A comprehensive, reliable and time-efficient pre-procedural planning which can be easily integrated in the intra-procedural workflow is of paramount importance, whether to delineate anatomical targets or to identify the optimal trajectory or path which avoids surrounding vital organs. In this research theme, we are developing novel approaches in statistical shape modeling and computational anatomy based on reinforced learning to classify, segment and register multimodal high-resolution diagnostic images.
Development of a 3D ultrasound elastography platform for in vivo wall strain analysis of abdominal aortic aneurysm
Winter 2020: Hongliang Li (Postdoc)
Abdominal aortic aneurysm (AAA) is an abnormal dilatation of the aorta in the abdomen due to a wall weakening caused by atherosclerosis. AAA rupture is the 13th cause of death in North America. While indications for a rupture intervention are based on AAA maximal diameter (5 cm), 23% of ruptured AAAs are less than 5 cm and in large AAAs, rupture rate could be lower than expected. This single indication would induce under or over estimation of rupture risk.
The project objective is to use 3-D US imaging to create a 3-D mapping of AAA strain and register the strain mapping on a single phase CT to improve the characterization of AAA vulnerability, which could be a new risk biomarker to predict AAA growth and rupture.
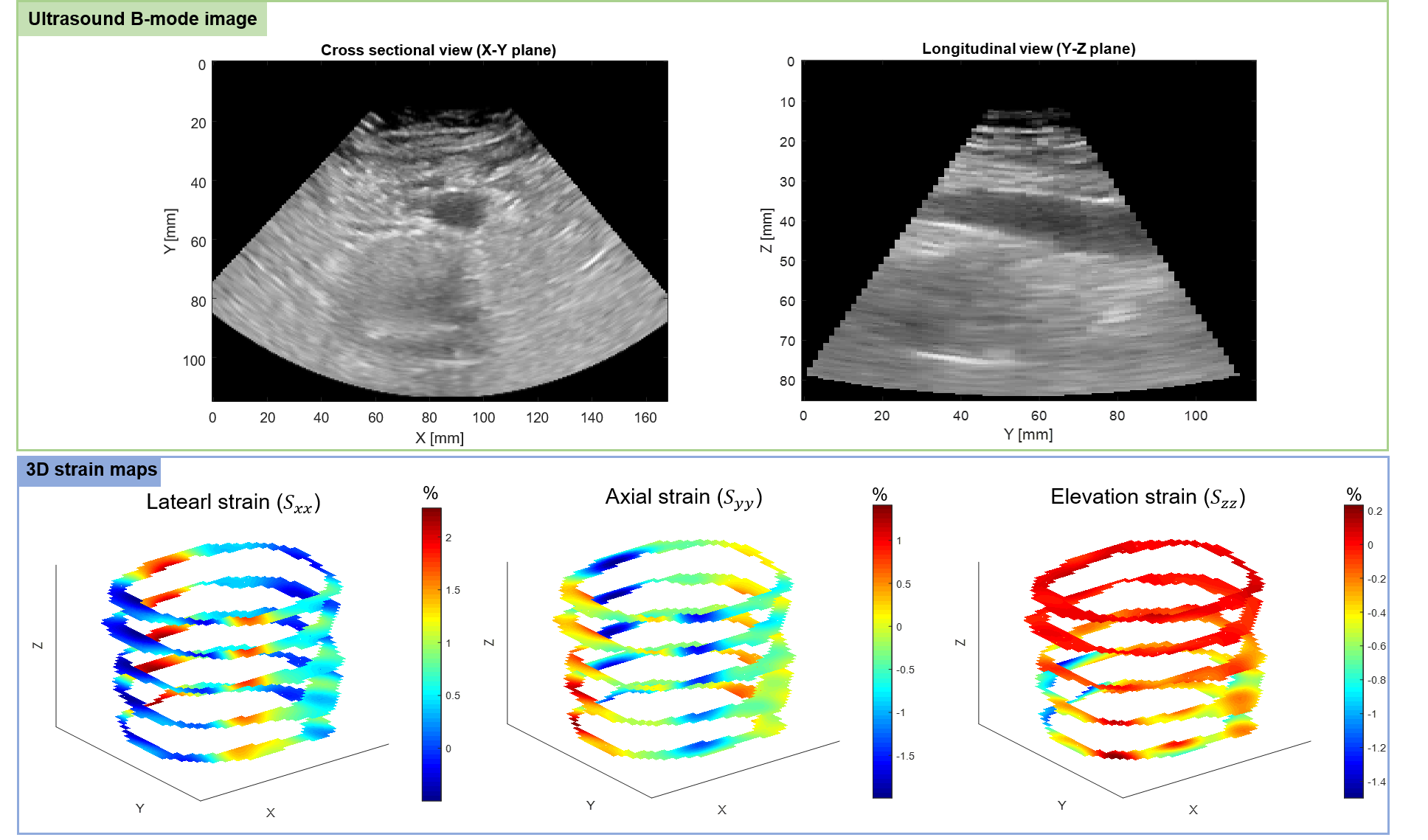
Submission: IUS 2020
Role of Quantitative Magnetic Resonance Biomarkers in Tumor-targeted Prostate Interventions
Stephanie Alley (Doctorate)
Multi-parametric magnetic resonance imaging (mpMRI) has proven to be an excellent tool in the diagnosis, staging, planning, treatment, and monitoring of prostate cancer because of its ability to depict soft-tissue contrast at high resolution. Despite advances in this area, discrepancies often occur between study findings due to a lack of reproducibility in mpMRI analyses. The goal of this work is to develop a pipeline of processing and analysis methods related to mpMRI of the prostate that will generate robust and reliable quantitative MR biomarkers that can be used to effectively target tumors interventionally.
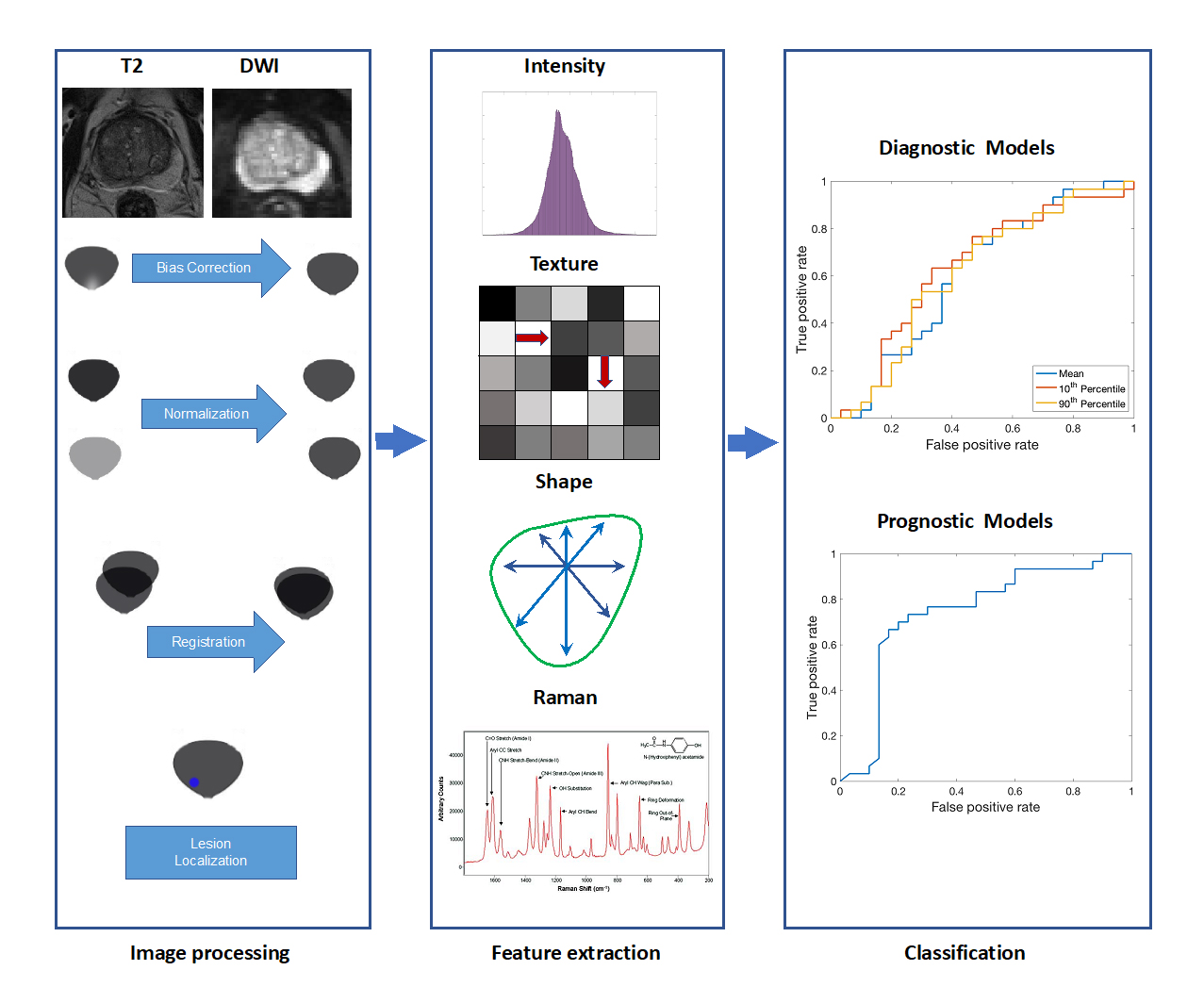
Address the multi-rater variability in manual lesion segmentation in medical images and its impact on deep learning segmentation models
Fall 2021: Zeinab Abboud (Doctorate)
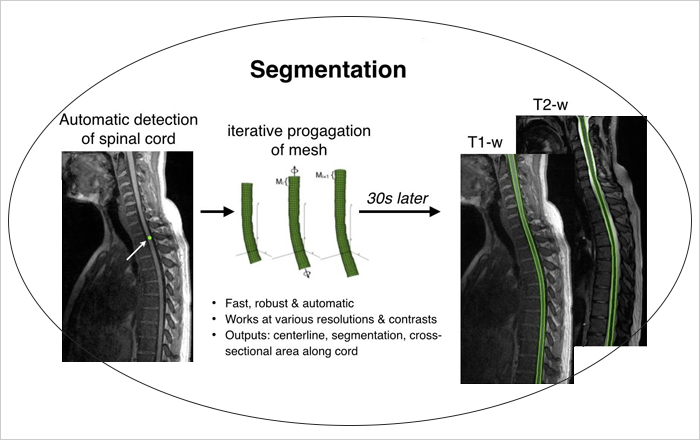
Spinal cord segmentation has a tremendous importance for various applications, from surgical planning to neurodegenerative diseases diagnosis/prognosis. However, only a few technics exist that segment the spinal cord automatically and for different type of MR contrasts. The objective of this project is to develop an automatic segmentation method for the spinal cord in MR images (T1- and T2-weighted) which manage the variable curvature of the spinal cord as well as contrast variation and artefacts. We will develop this method based on the multi-resolution propagation of deformable models, coupled with local contrast adaptive mechanisms. The automation will be provided by an independent spinal cord detection module, giving an approximation of spinal cord position and orientation in the volume. Coupled with an automatic vertebral level identification algorithm, this method will provide quantitative metrics of the spinal as cross-sectional areas and volumes in a generic vertebral coordinate system, allowing easy inter- and intra-patient comparisons for large-cohort studies. This developed method will be freely available onhttp://sourceforge.net/projects/spinalcordtoolbox/.
Benign epilepsy with centrotemporal spikes (BECTS) is one of the most common childhood epilepsy syndromes, which occurs in children aged from three to 13 years old. Initially, BECTS was considered as benign, but recently some studies have found cognitive and behavioral deficits, which may persist even after remission. Recent neuroimaging studies have found a link between these cognitive deficits and dysfunctions in specific brain structures, which shows the possibility of neuroanatomical alterations in these brain regions. In this research project, we aim to propose an automatic morphological analysis framework in BECTS to detect the subtle neuroanatomical alterations in children with BECTS, compared to normal controls. To this end, we develop an iterative group-wise coregistration and cosegmentation process, which enables automatic segmentation of sub-cortical structures in multiple images. Then, we design a framework for matching 3D sub-cortical surface meshes and investigating the group-wise structural differences between two populations of surfaces, i.e., healthy and pathological subjects. Finally, we propose a methodology to assess the association between morphological alterations and cognition.

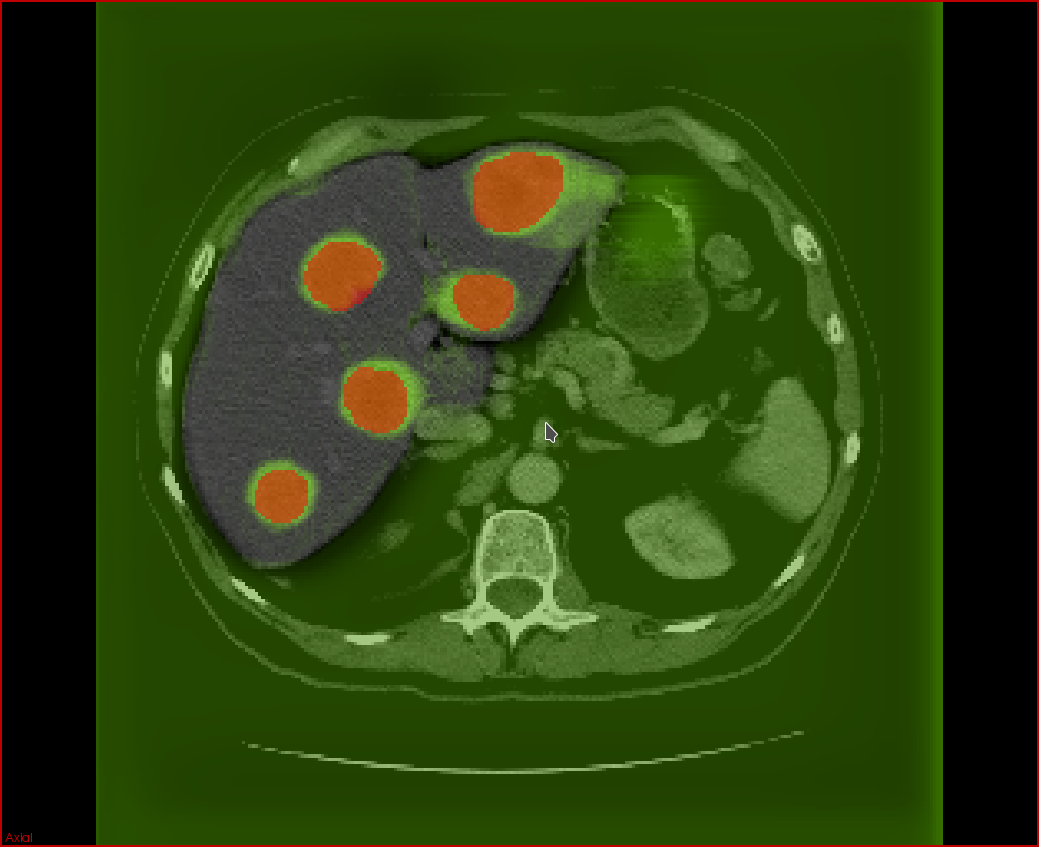
Secondary liver tumors (metastases), in aggragate with primary liver tumors (hepatocellular carcinoma), are the most common cause of cancer-related mortality in North America. Currently, there is insufficient automation in the processing of 3D tumor scans from imaging modalities such as computed tomography (CT) and magnetic resonance imaging (MRI). In particular, there is much room for the development of automated tissue classification, lesion detection, and lesion and organ segmentation. In the context of cancer diagnosis, such tools are useful for the improvement of computer-aided diagnoses (CADx) of cancer tumors.
The aim of the project is to help radiologists reduce the rate of false negatives as well as the time required to offer an accurate diagnosis and follow-up. The goal is to develop the tools based on deep learning techniques for a cancer CADx software that can be seamlessly integrated within an existing PACS (Picture Archiving Communication System) and RIS (Radiology Information System).
Automatic segmentation of lesions will allow accurate estimation of tumor volume and perfusion, aiding in the estimation of tumor burden and in the evaluation of response criteria for solid tumors. Currently, these evaluations are obtained using 2D measurements, such as the length and width of tumors. Deep neural network models require significant amounts of data to train well-performing models which is a challenge due to the scarcity of subjects that may exhibit tumor types of interest and the difficulty in labeling large numbers of medical images for training. Using manually labeled data from clinical collaborators, we plan to develop models that will detect and segment tumors in a liver segmented from CT and MRI images. The development of these models will open the prospect for improved accuracy at reduced variability with minimal human intervention. This has the potential to expand the use of medical imaging for tumor screening, diagnosis, and followup, ultimately improving patient care.

MRI is the standard medical imaging method to detect neurological problem with brain images. For neonates, new ultrasound probes can acquire 3D images quasi instantly and are easy to use. Since, MRI and 3D US images were both acquired for each patients, a study is realize at first to compare US and MRI brain volume as well as ventricles volume. Then, an image registration is realize between 3D US images and an MRI atlas in order to match the regions of interest. After a segmentation is realize in order to extract the ventricles or other regions of interest. This approach allow us to extract 3D volumes from brain with US images instead of MRI, diminishing times and complexity.

In clinical routine, manual segmentation techniques are applied upon CT-images for the diagnosis of the liver tumors. Recently many researches have been done for the automated segmentation methods by using artificial intelligence. However, prediction and segmentation of liver lesions from follow-up scans remains still an open problem because of some challenges such as tumors evolution and lesions growth. The goal of our project is to predict these changes from follow-up scans by using the latest developments in artificial intelligence and deep learning.
The segmentation of cancerous liver tumors from abdominal MRI or CT images is important for both the diagnosis and treatment of liver cancer. Currently, this segmentation is performed manually; however, manual segmentation is time consuming and requires expert knowledge. Automating the segmentation process is an area of active research and available methods do not achieve adequate performance. The objective of this project is to develop a model for the challenging task of automatically segmenting cancerous liver tumors from MRI images. Liver tumor segmentation is particularly challenging due to highly variable size, shape, intensity and texture. This variability will be incorporated into the model through machine learning. Another challenge is the small amount of available training data due to the small size of available datasets for a given imaging system. Taking these challenges into consideration, an appropriate learning-based classifier will be developed and combined with a novel deformable surface model to refine the resulting segmentation.

The key point for radiotherapy treatment success is to target precisely the tumors while preserving healthy tissue from ionizing radiation. This requires precise calibration of the ionizing rays which is usually performed using pre-operative data. The pre-operative images of the patient anatomy are registered to the intraoperative patient position, allowing the radiotherapy system to know the tumor location. However, during a radiotherapy treatment the patient is conscious and free to breath. Thus the tumors move and their positions do not correspond to the ones given by the registered pre-operative images.
The idea of the project is to predict the tumor motion inside the liver to adjust the radiation beam in real time during the treatment in order to maximize tumor exposition and to minimize healthy tissue damages.
We propose to construct a population based motion model from dynamic MR images of the abdomen under free breathing. This model will then be personalized to the patient data using machine learning techniques.
Registration of pre-operational MRI with C-arm fluoroscopic X-Ray is a challenging process. Recent advancements in deep learning, with Generative Adversarial Networks (GANs), have shown promising results in the synthesis of CT images of the brain given their pre-op MRI's. Our end-goal is the automate the MRI/C-arm fluoroscopy registration of vertebrae with CT imaging as a stepping stone; given that automatic CT/C-arm fluoroscopy registration is more doable. We will first train a GAN to predict CT images of vertebrae given their MRIs and then automatically register the synthetic CT with the C-Arm fluoroscopy. The same transformation will then be applied to the initial MRI to obtain a final MRI/C-Arm fluoroscopy registration.
The motion and deformation of the liver during free-breathing is an important problem during tumor targeting procedures such as radiotherapy: in order to maximize neoplastic tissue exposition and to minimize damage to healthy tissues, the focus point of the radiation beam must follow the tumor position. However, during this type of treatment, the patient is free to breath and thus the respiratory motion needs to be compensated.
The goal of this project is to construct a motion model of the liver during breathing that takes into account inter-cycle variations to predict the tumor position during radiotherapy and allow for better targeting of the tumor. The method relies on using deep generative networks to learn the organ motion from a population dataset.
Hepatocellular carcinoma (HCC) is the sixth most common cancer, the second leading cause of cancer death in the world, and its incidence is still increasing in North America. Trans-arterial chemoembolization (TACE) is an accepted treatment for non-surgical liver cancer. Recently, four-dimensional (4D) flow MRI has become an important tool to study blood velocity and flow in different vascular territories and allows a posteriori blood flow quantification throughout a 3D field of view. 4D flow MRI analysis could thus be of potential benefit to complement the pre-TACE evaluation of HCC patients not eligible for surgery and improve the guidance of endovascular treatments.
The goal is to study and evaluate the feasibility of 4D flow MRI measurements in hepatic arteries of patients with hepatocellular carcinoma (HCC) before and after transarterial chemoembolization (TACE) treatment.


